As the rules for cell permeability continue to be elucidated, peptides are increasingly being used to deliver either themselves or cargo to the cell’s interior. One thing is clear, increasing the overall cationic charge of the peptide enhances it’s delivery to not only the cytoplasm, but also the nucleus or other subcellular compartments. To achieve the positive charge, large numbers of arginine residues are most often incorporated into the peptide sequence.
This begs the question though, should I change my cleavage protocol? In today’s post, I’ll evaluate several lengths of time used to cleave and fully deprotect an Arg-rich peptide sequence.
Certainly synthesizing peptides rich in arginine can be a challenge, the challenge is further extended in the cleavage reaction. While the physical removal of a peptide from the resin is often rapid, it is well established that removing the Arg Pbf-protecting group can be more challenging. Incorporating multiple arginine residues into a peptide, particularly if they are close in linear sequence, can further compound this issue. I, and I’m sure many others of you out there, have purified a crude peptide only to find that a significant portion of the crude material contains the desired peptide with residual Pbf protecting group(s).
So to figure this out, I synthesized a large batch of the TAT cell-penetrating peptide (GRKKRRQRRR-NH2) and divided the resin into 3 roughly equal portions. I then suspended the dry resin in cleavage cocktail (95:2.5:2.5 TFA:water:TIS) and stirred for 1, 2, or 3 hours. I chose to use a minimal volume of cleavage cocktail to reduce the amount of TFA handling after the cleavage was completed.
I decided to quickly analyze each of the cleaved peptide samples by directly injecting the crude mixture, dissolved in 1% HFBAaq, onto my Biotage® Isolera Dalton 2000. I did expect to see a significant number of masses, given that this peptide will ion pair with any negatively charged counterion to a significant extent, and was not disappointed. After some deconvolution though, I identified both my desired peptide and a des-Arg deletion sequence in the mixture subjected to cleavage for a full three hours, Figure 1.
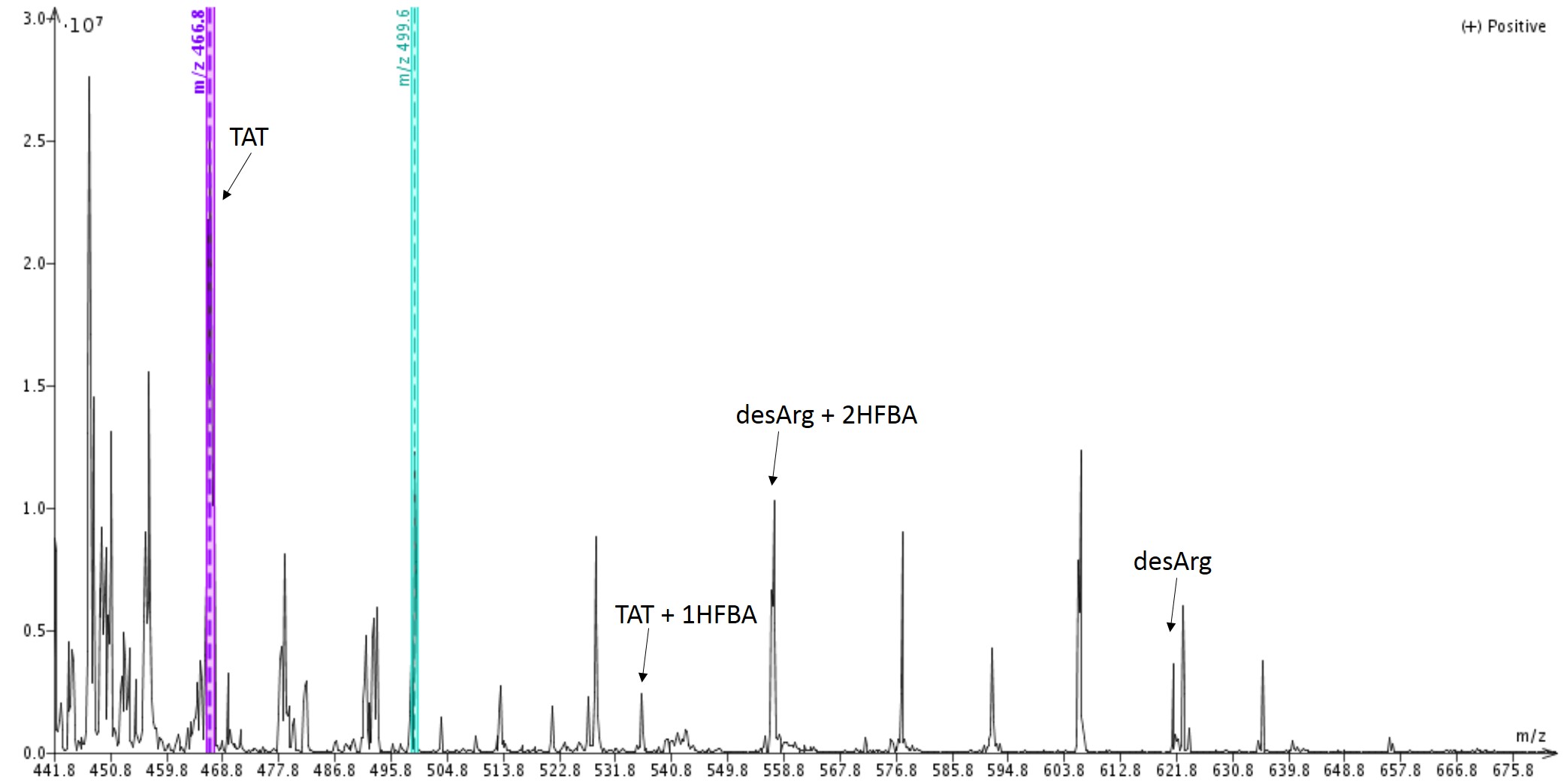
Figure 1: Mass spectrum of crude TAT peptide stirred with cleavage cocktail for 3 hours. Masses for Tat peptide and the des-Arg deletion sequence are highlighted.
When I evaluated the samples cleaved for only one hour, I identified peptide masses that correspond to the desired TAT sequence, but also masses corresponding to TAT with a residual Pbf protecting group. Figure 2.
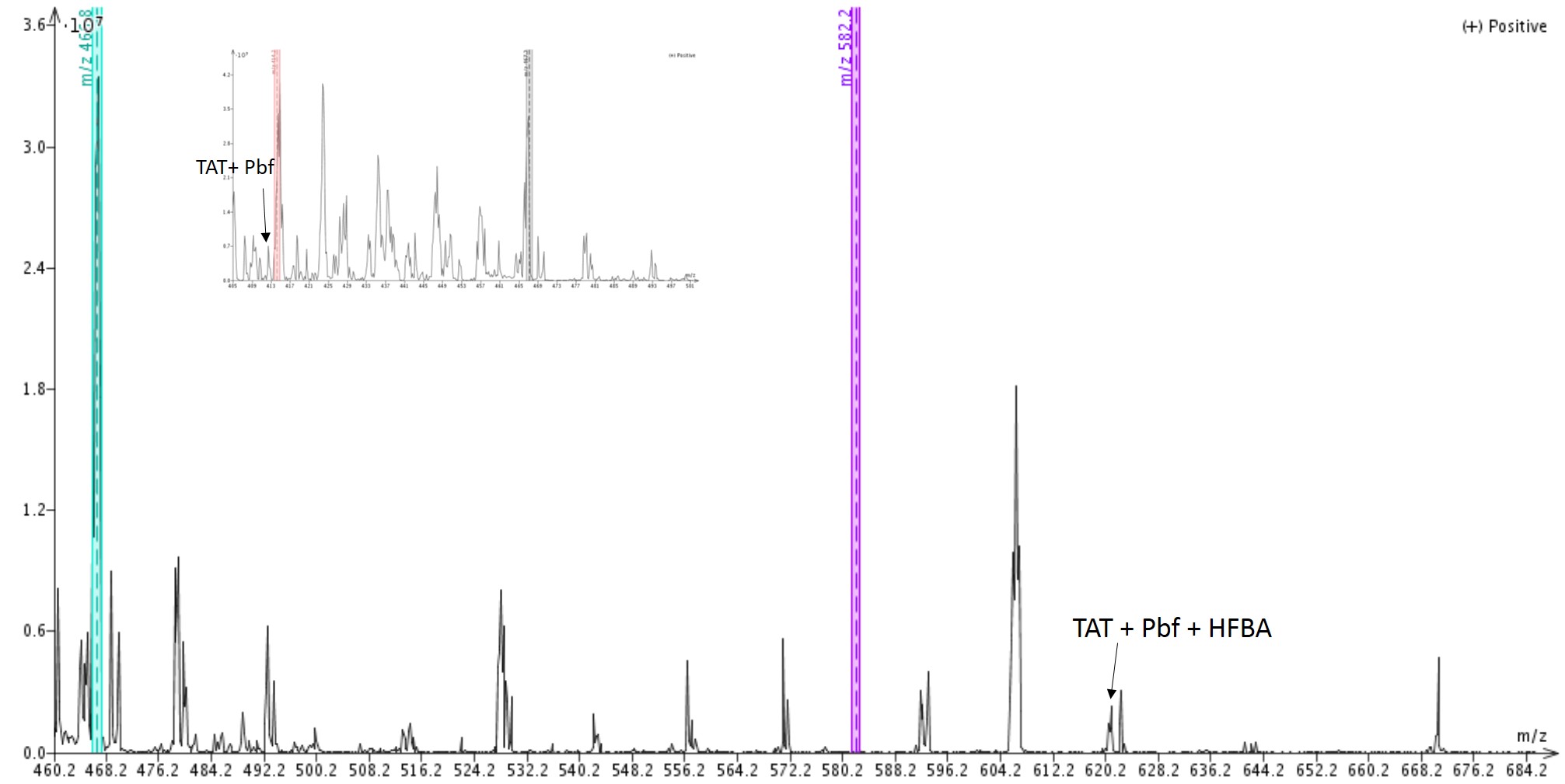
Figure 2. Mass spectrum of crude Tat peptide cleaved for only 1 hour. Masses assigned to peptides still containing a Pbf protecting group are indicated.
Gratifyingly, the residual protecting group signal decreases below the level of detection as a function of cleavage reaction time (data not shown) as expected. I would have liked to see the parent peptide ion intensities grow, but using ion counts to measure concentration is not recommended, especially without an internal standard.
You may notice however, that a molecular weight of 621 has been indicated and assigned to two different peptides. I have made the assigment after careful consideration of all three spectra. Interestingly, this mass is completely absent from the peptide cleaved for two hours. This suggests and equilibrium of sorts, where after only a short cleavage time residual Pbf-protected Tat peptide remains, and after a longer reaction time the unprotected des-Arg sequence is detected. If this mass represented a single species, I would have expected a consistent ion count across all injections (in the case of des-Arg) or a complete loss of signal all together (for protected Tat).
With this information in hand, I’d recommend increasing your room temperature cleavage reaction time to a full three hours when cleaving peptides rich in arginine residues. An interesting alternative experiment is to increase the total cleavage cocktail volume or alter the additive composition. Maybe I’ll try those next!
To learn more about Biotage tools designed to improve your peptide workflow efficiency - delivering a pure peptide product more quickly, follow the link below.

 Organic Workflow
Organic Workflow Peptide Workflow
Peptide Workflow Scale-Up Flash Purification
Scale-Up Flash Purification  Sample Preparation
Sample Preparation Biomolecule Purification
Biomolecule Purification Oligo synthesis
Oligo synthesis Scavengers and Reagents
Scavengers and Reagents Service & Support
Service & Support Accessories & Spare parts
Accessories & Spare parts Investors
Investors Reports & News
Reports & News The Share
The Share Corporate Governance
Corporate Governance Calendar
Calendar Sustainability
Sustainability Our Offering
Our Offering Our History
Our History Our Locations
Our Locations Leadership
Leadership